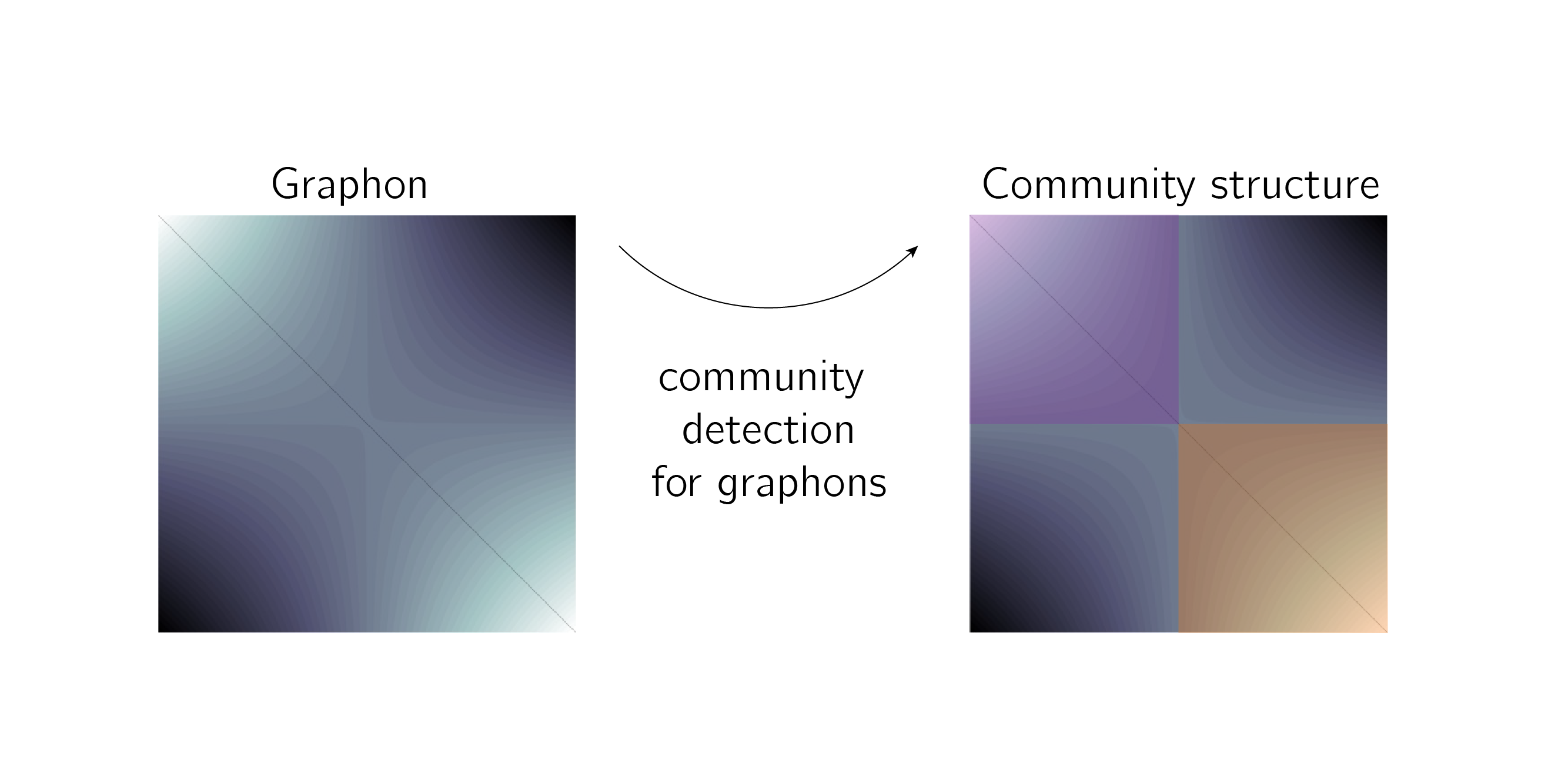
As used for "Modularity maximisation for graphons", please find code here.
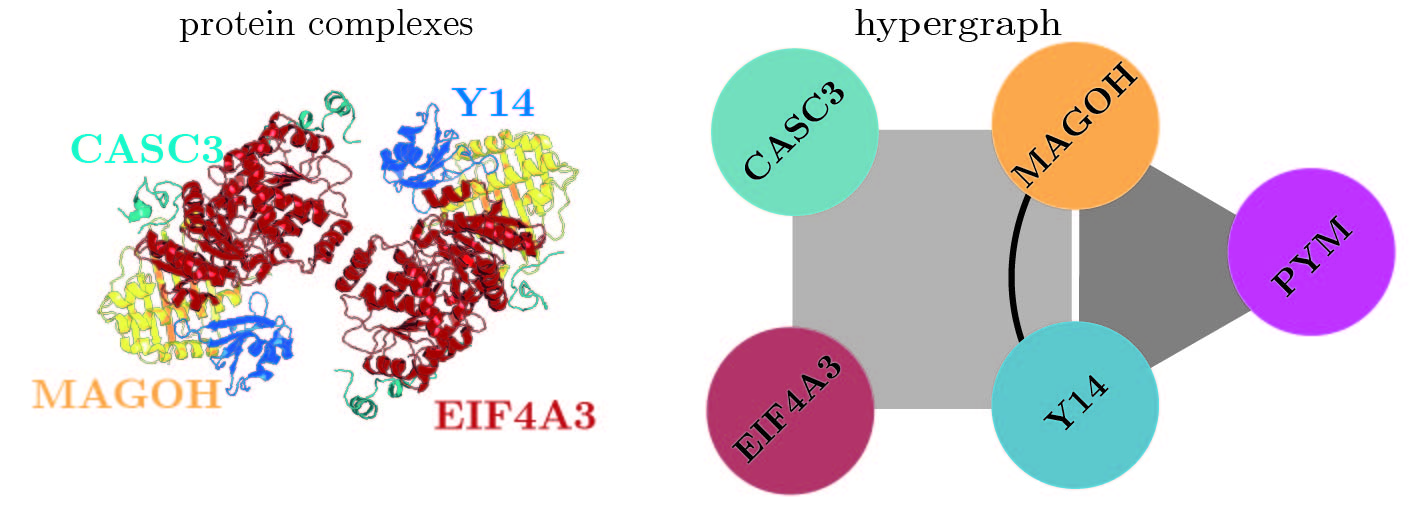
As used for "Hypergraphs for predicting essential genes using multiprotein complex data", please find code here.
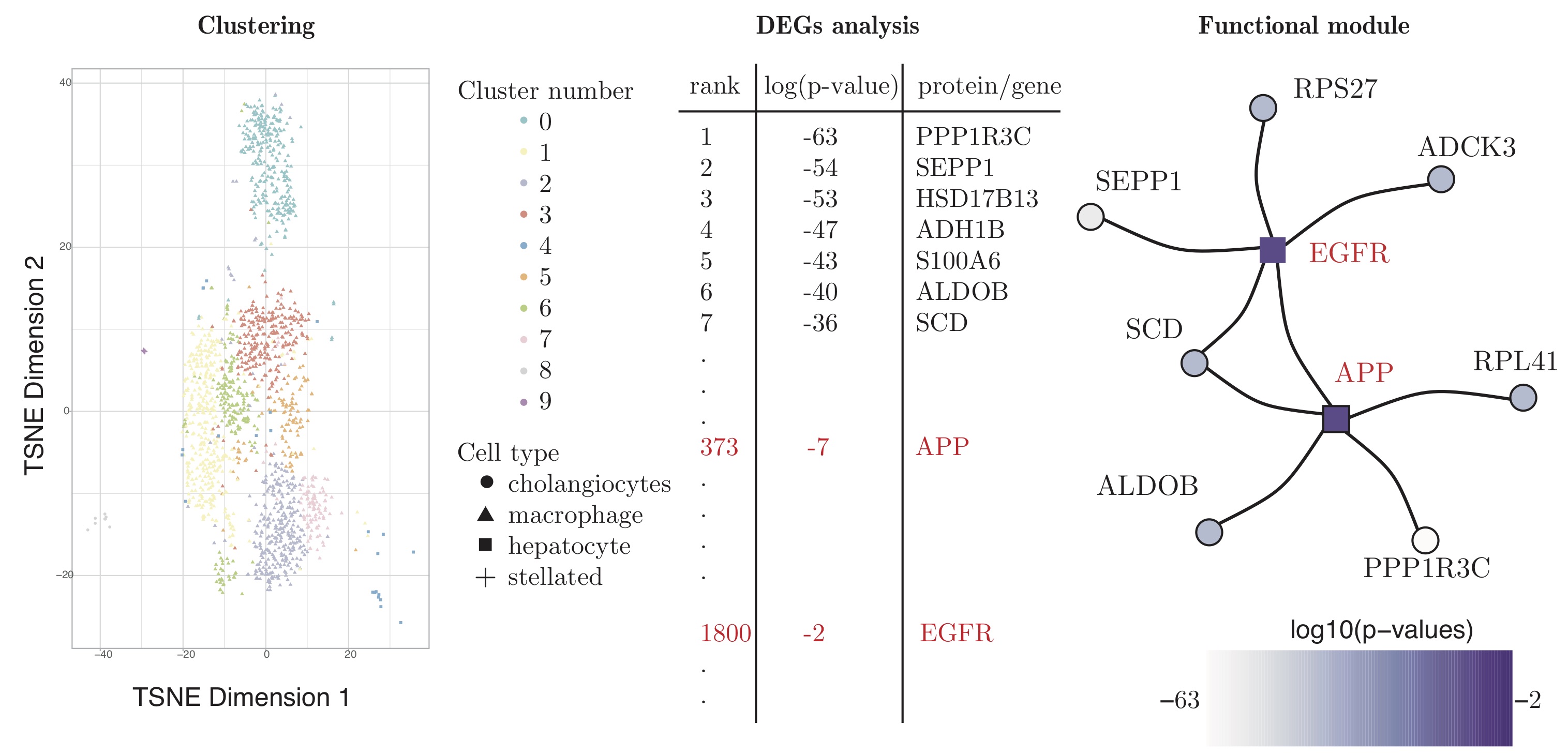
As used for "Functional module detection through integration of single-cell RNA sequencing data with protein–protein interaction networks", please find code here.
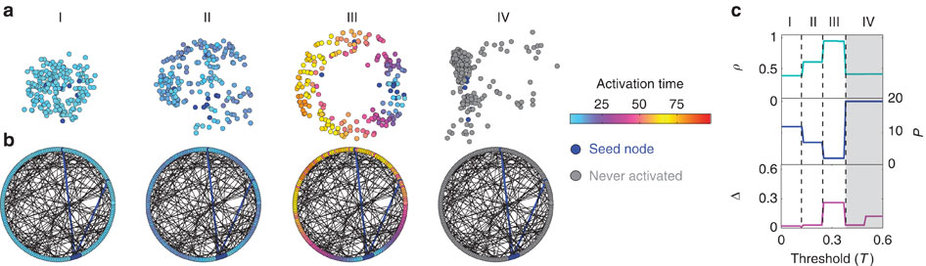
Benjamin F. Maier and I tought a two-week summer school for high-school students and the teaching material is available on GitHub.
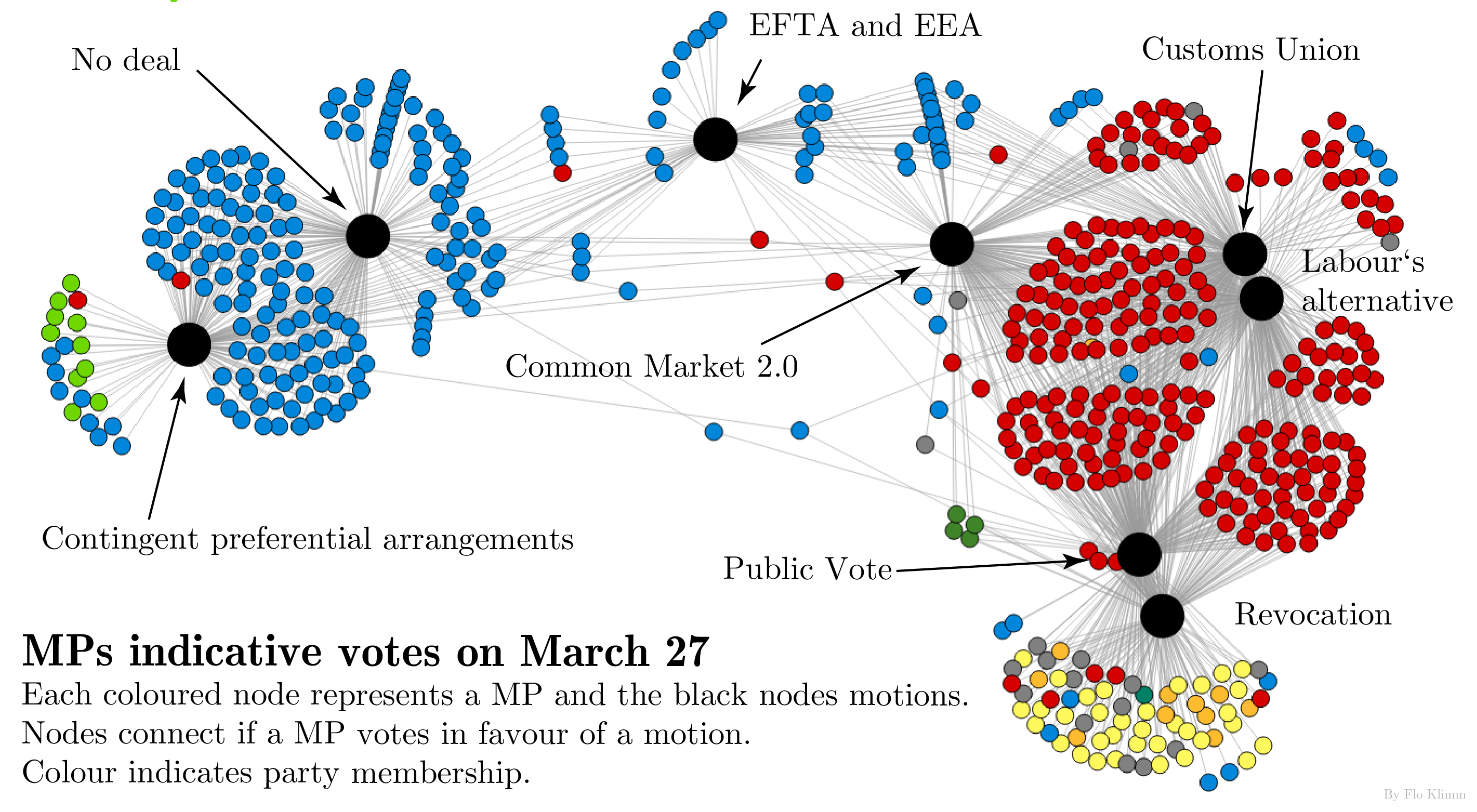
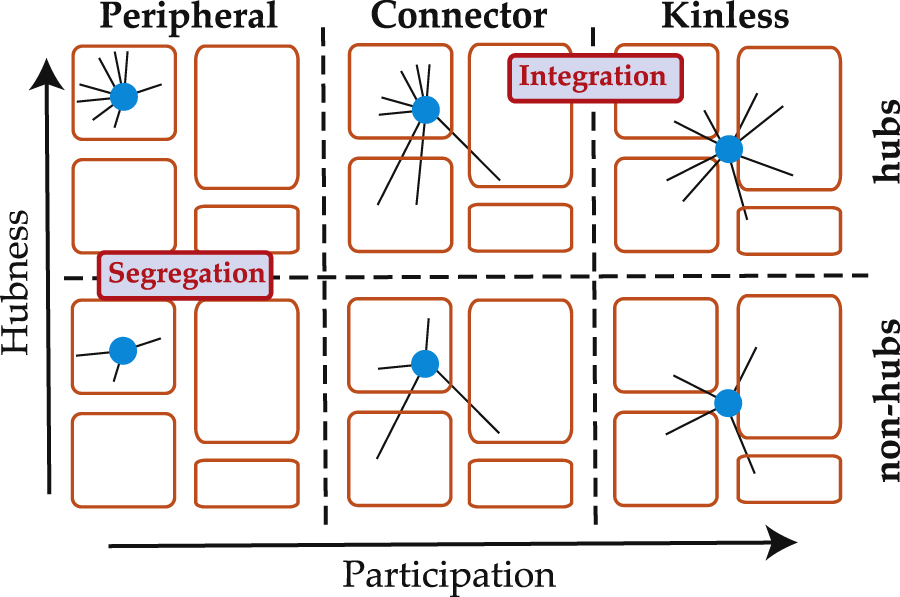
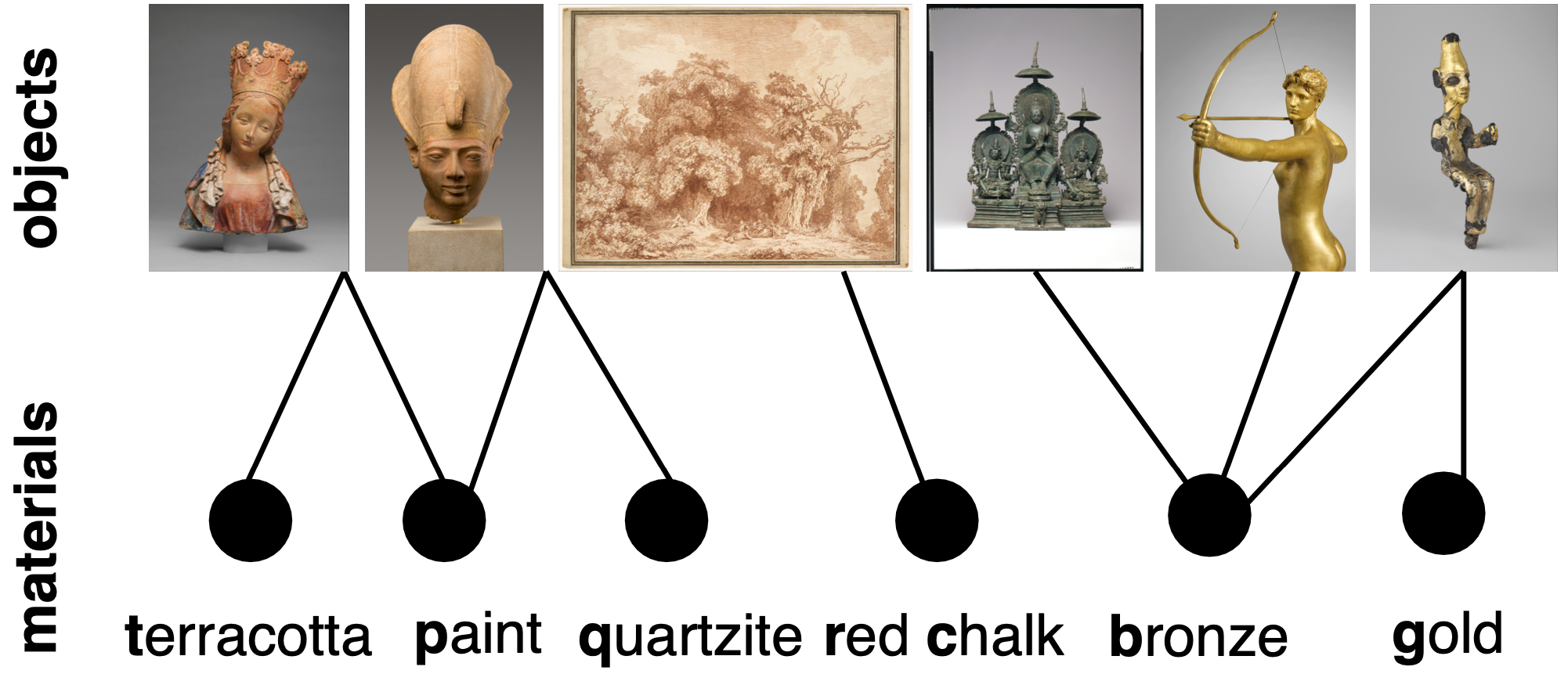
Data of the material -- object network from The Metropolitan Museum of Art. The Met kindly provides the updated raw data here. We constructed a network from the data in Fall 2014. Used in Griffin and Klimm, Oxford Handbook of Archaeological Network Research (Oxford University Press).
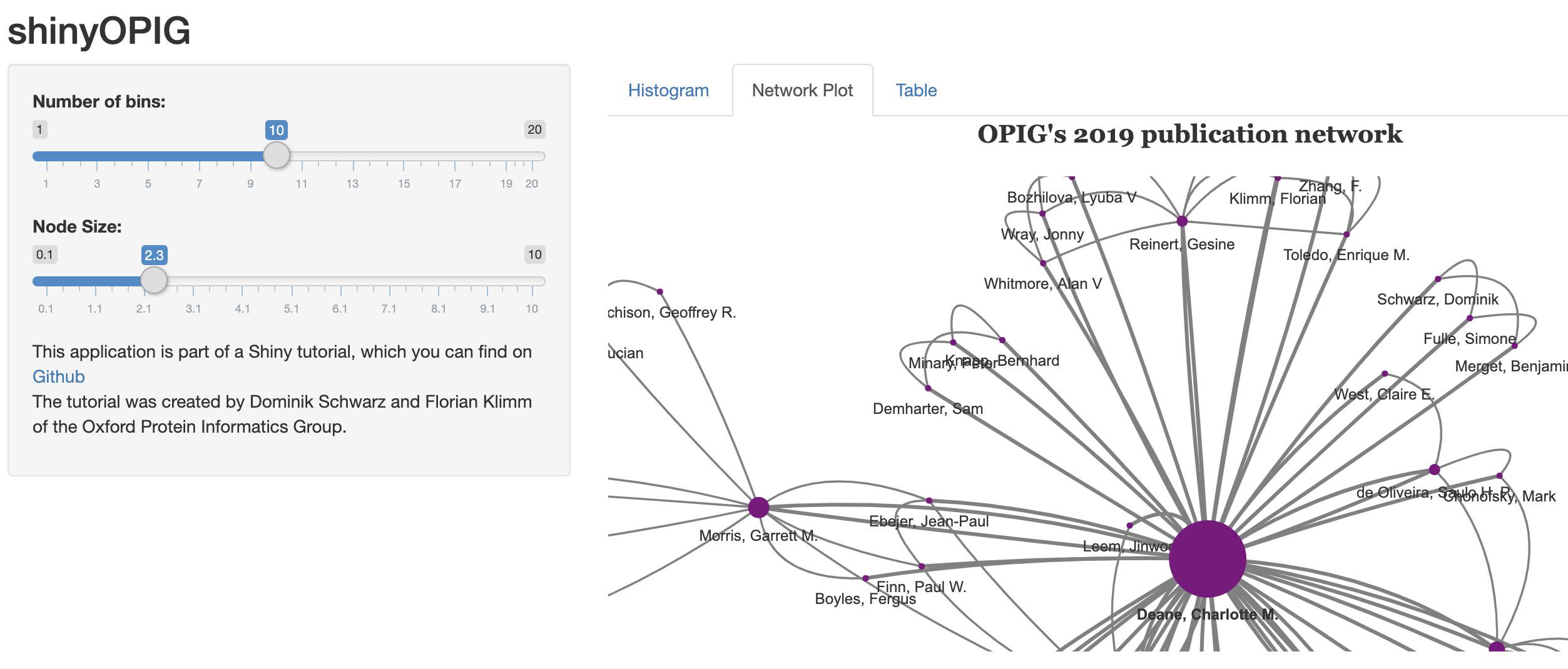
This tutorial illustrates the usage of the R libraries Shiny and visNetwork for the creation of interactive web apps. The tutorial is available here and you will learn to create this web app.
This excellent library is not created by myself but Jürgen Hackl. You can find it here. It is particularly well-suited for multilayer networks.